From Massachusetts General Hospital 03/11/23

The brain is made up of numerous types of cells that are organized into different structures and regions.
Although several important steps have been made towards building models of the human brain, the advances have not produced undistorted 3D images of cellular architecture that are needed to build accurate and detailed models.
In new research published in Science Advances, a team led by investigators at Massachusetts General Hospital (MGH), a founding member of Mass General Brigham (MGB), has overcome this challenge to offer scientists and clinicians a comprehensive cellular atlas of a part of the human brain known as Broca’s area, with detailed resolution to study brain function and health.
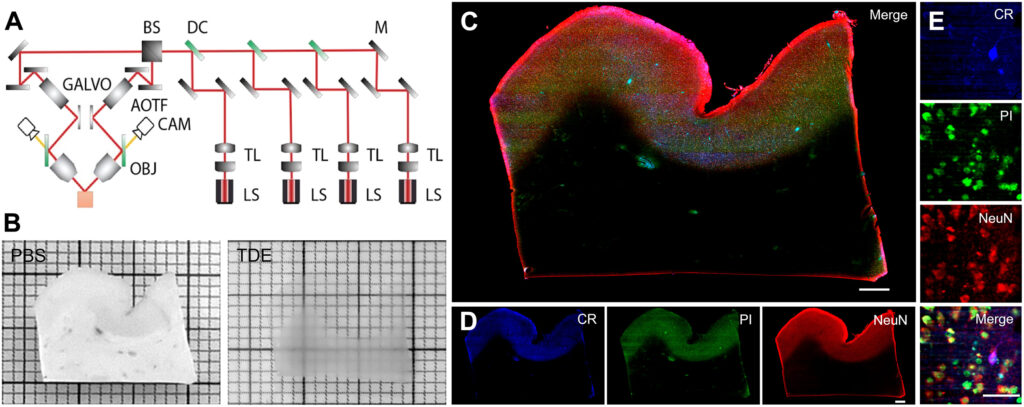
By combining different sophisticated imaging techniques—including magnetic resonance imaging, optical coherence tomography, and light-sheet fluorescence microscopy—the researchers were able to overcome the limitations associated with any single method to create a high-resolution cell census atlas of a specific region of the human cerebral cortex, or the outer layer of the brain’s surface.
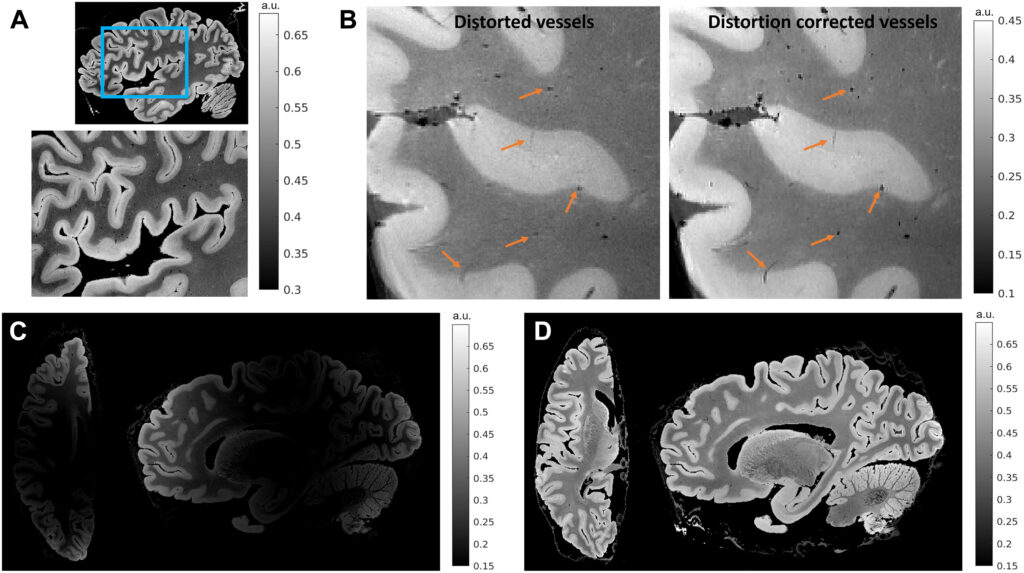
The team created such an atlas for a human postmortem specimen and integrated it within a whole-brain reference atlas.
“We built the technology needed to integrate information across many orders of magnitude in spatial scale from images in which pixels are a few microns to those that image the entire brain,” says co–senior author Bruce Fischl, PhD, director of the Computational Core at the Athinoula A. Martinos Center for Biomedical Imaging at MGH and a professor in Radiology at Harvard Medical School.
Ultimately, the methods in this study could be used to reconstruct undistorted 3D cellular models of particular brain areas as well as of the whole human brain, enabling investigators to assess variability between individuals and within a single individual over time.
This will provide insights into the presence and spread of pathologic changes that occur in neurodegenerative and psychiatric illnesses.
“These advances will help us understand the mesoscopic structure of the human brain that we know little about. Structures that are too large and geometrically complicated to be analyzed by looking at 2D slices on the stage of a standard microscope, but too small to see routinely in living human brains,” says Fischl.
“Currently we don’t have rigorous normative standards for brain structure at this spatial scale, making it difficult to quantify the effects of disorders that may impact it such as epilepsy, autism, and Alzheimer’s disease.”
light-sheet fluorescence microscope



