From University of Nottingham 17/11/23

A UK-based team of Scientists, led by experts from the University of Nottingham and Imperial College London, have completed construction of a synthetic chromosome as part of a major international project to build the world’s first synthetic yeast genome.
The work, which is published in Cell Genomics, represents completion of one of the 16 chromosomes of the yeast genome by the UK team, which is part of the biggest project ever in synthetic biology; the international synthetic yeast genome collaboration.
The collaboration, known as ‘Sc2.0’ has been a 15-year project involving teams from around the world (UK, US, China, Singapore, UK, France and Australia), working together to make synthetic versions of all of yeast’s chromosomes.
Alongside this paper, another 9 publications are also released today from other teams describing their synthetic chromosomes.
The final completion of the genome project – the largest synthetic genome ever – is expected next year.
This effort is the first to build a synthetic genome of a eukaryote – a living organism with a nucleus, such as animals, plants and fungi.
Yeast was the organism of choice for the project as it has a relatively compact genome and has the innate ability to stitch DNA together, allowing the researchers to build synthetic chromosomes within the yeast cells.
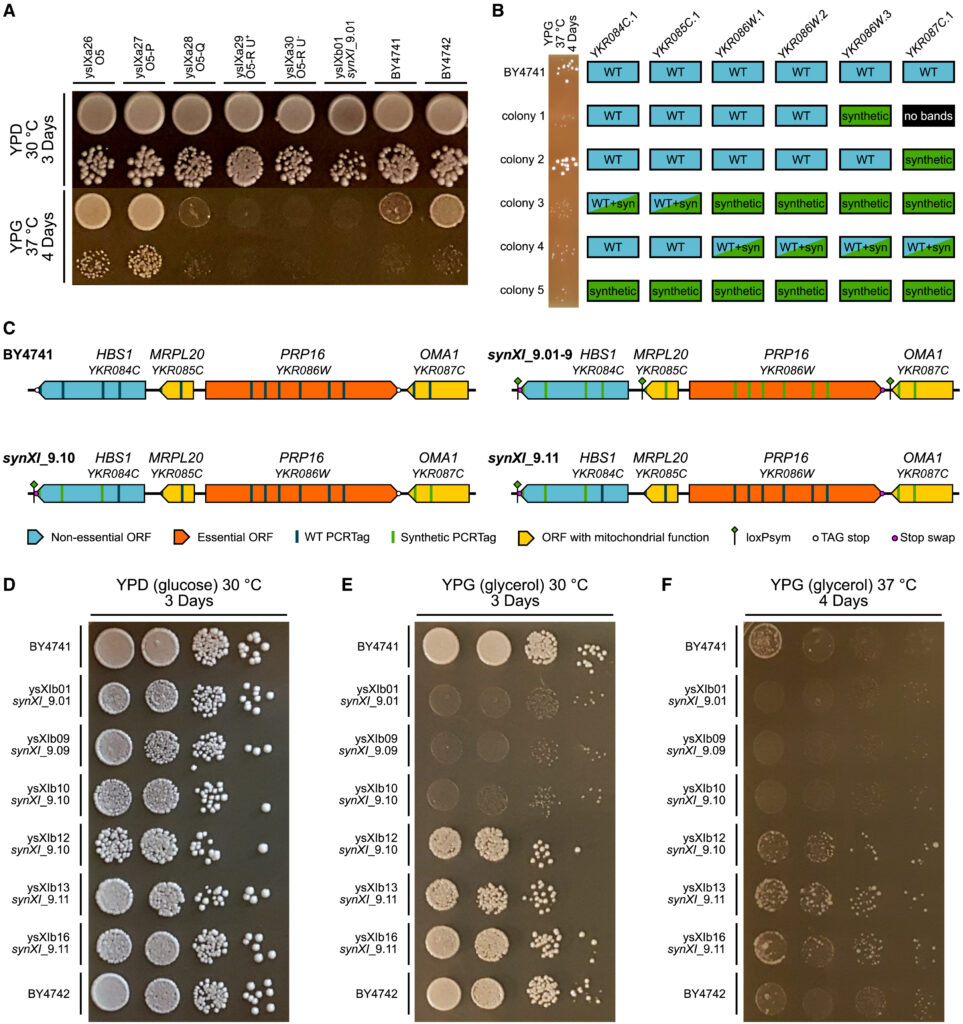
(A) Growth spot assays of synXI assembly intermediates following megachunk integration on YPD (glucose) and YPG (glycerol) to test respiratory function. For each strain and condition, the top spot is a ×10−1 dilution, and the bottom spot is a ×10−3 dilution.
(B) PCRTag screening results at the YKR084C.1-YKR087C.1 locus for debugging megachunk Q transformant colonies, with corresponding ×10−3 dilution YPG 37°C growth spots on the left, indicating respiratory function.
(C) Schematics of the HBS1-OMA1 locus in BY4741 and the in vivo synXI iterations.
(D–F) Growth spot assays with strains involved in respiratory growth defect debugging on YPD (glucose) and YPG (glycerol) to test respiratory function. Dilution spots increase in steps of ×10−1 from ×100 on the left to ×10−3 on the right. Credit: Cell
Humans have a long history with yeast, having domesticated it for baking and brewing over thousands of years and, more recently, using it for chemical production and as a model organism for how our own cells work.
This relationship means that we know more about the genetics of yeast than any other organism.
These factors made yeast the obvious candidate.
The UK-based team, led by Dr Ben Blount from the University of Nottingham and Professor Tom Ellis at Imperial College London, have now reported completion of their chromosome, synthetic chromosome XI.
The project to build the chromosome has taken 10 years and the DNA sequence constructed consists of around 660,000 base pairs – which are the ‘letters’ making up the DNA code.
The synthetic chromosome has replaced one of the natural chromosomes of a yeast cell and, after a painstaking debugging process, now allows the cell to grow with the same fitness level as a natural cell.
The synthetic genome will not only help scientists to understand how genomes function, but it will have many applications.
Rather than being a straight copy of the natural genome, the Sc2.0 synthetic genome has been designed with new features that give cells novel abilities not found in nature.
One of these features allows researchers to force the cells to shuffle their gene content, creating millions of different versions of the cells with different characteristics.
Individuals can then be picked with improved properties for a wide range of applications in medicine, bioenergy and biotechnology.
The process is effectively a form of super-charged evolution.
The team has also shown that its chromosome can be repurposed as a new system to study extrachromosomal circular DNAs (eccDNAs).
These are free-floating DNA circles that have “looped out” of the genome and are being increasingly recognised as factors in ageing and as a cause of malignant growth and chemotherapeutic drug resistance in many cancers, including glioblastoma brain tumours.
Dr Ben Blount, one of the lead scientists on the project, is an Assistant Professor in the School of Life Sciences at the University of Nottingham.
He said: “The synthetic chromosomes are massive technical achievements in their own right, but will also open up a huge range of new abilities for how we study and apply biology.”
“This could range from creating new microbial strains for greener bioproduction, through to helping us understand and combat disease.”
“The synthetic yeast genome project is a fantastic example of science on a large scale that has been achieved by a large group of researchers from around the world.”
“It’s been a great experience to be part of such a monumental effort, where all involved were striving towards the same shared goal.”
Professor Tom Ellis from the Centre for Synthetic Biology and Department of Bioengineering at Imperial College London, said: “By constructing a redesigned chromosome from telomere to telomere, and showing it can replace a natural chromosome just fine, our team’s work establishes the foundations for designing and making synthetic chromosomes and even genomes for complex organisms like plants and animals.”